Multi-group analysis
Note: This step is optional and the default option is to select all the available samples for analysis. One may wish, however, to restrict analysis to only a subset of all the available samples for an experiment. We have described earlier how to generate a two-group sample differential expression signature by clicking "Create a Signature" (see "Create a Signature" link on the left of this help file). The following section will explain how to find differentially expressed genes between multiple groups of samples.
To find differentially expressed genes between multiple groups of samples within a dataset, we must click "Multi-group Analysis" link on the top right corner from the dataset landing page as shown in the figure below.
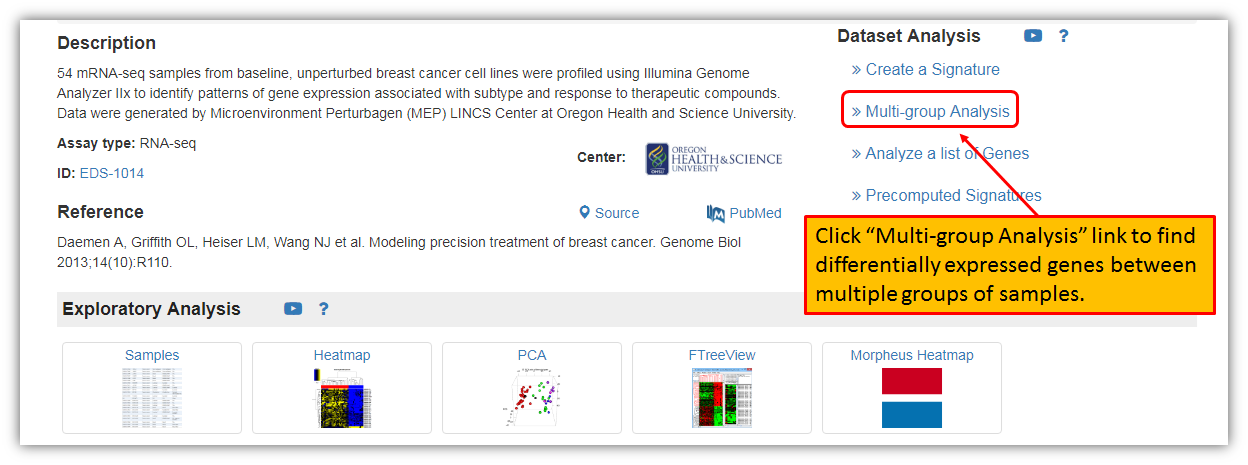
The figure below shows a landing page for Multi-group analysis, i.e. finding differentially expressed genes between multiple groups of samples within selected dataset. Each dataset will have all of its samples listed in different subgroups. This screen is allowing you to select the grouping variable and to specify clustering type. For this example, let's select grouping variable for the analysis based on the CK14 status and to include all of the samples within a dataset (there are 3 groups for CK14 status: group with no information given about CK14, one group with a negative CK14 and one group with positive CK14). The default clustering type is "genes", but you may change it (other option are in drop down menu: none, genes, samples, samples and genes).
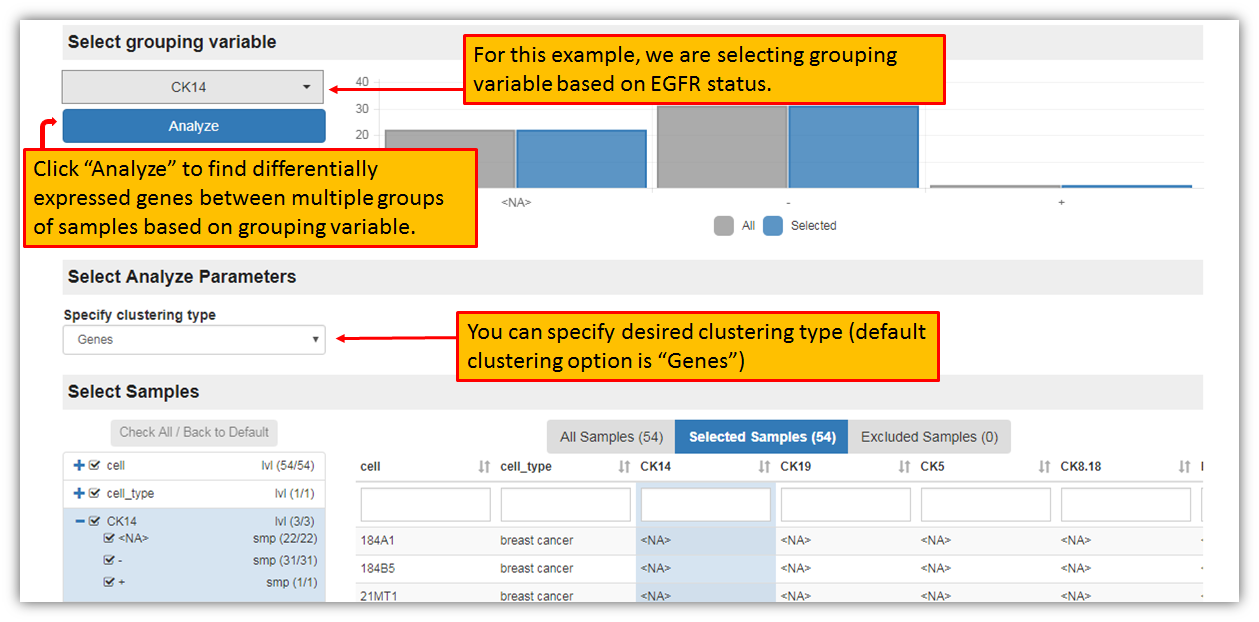
After the selection of sample grouping variable, specifying clustering type and clicking "Analyze", iLINCS portal will generate signature and will take you to generated signature landing page, where you may further analyze, view and download the signature as seen in the figure below.
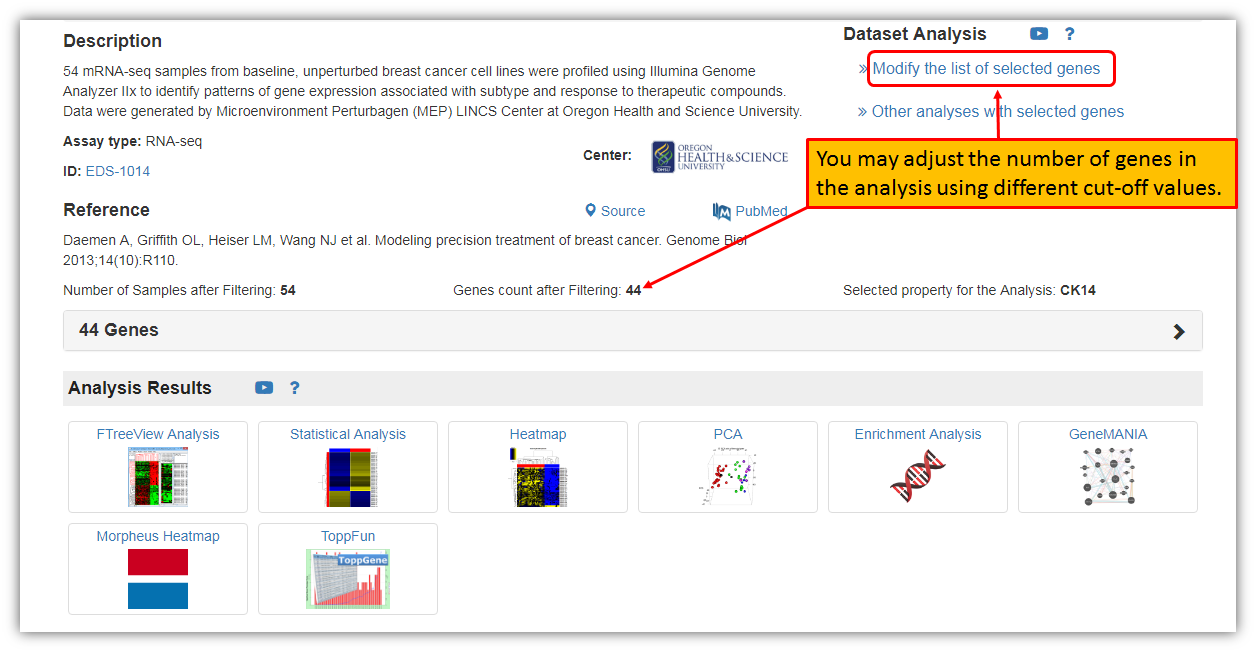
Just as before, you have the option to change/adjust the number of genes in the analysis using Shiny Volcano plot by selecting "Modify the list of selected genes" by using different Log2 Fold Change and P-value cut-offs.
Created with the Personal Edition of HelpNDoc: Free help authoring tool