Start by exploring Signatures
In the "Signatures" pipeline, you may explore, analyze and visualize over 300,000 pre-computed LINCS signatures (i.e. list of "scores" (activity levels) for a list of genes or for all genes in the genome "genome-wide signatures"). One would land on the Signatures landing page by clicking "Signatures" on the iLINCS portal header.
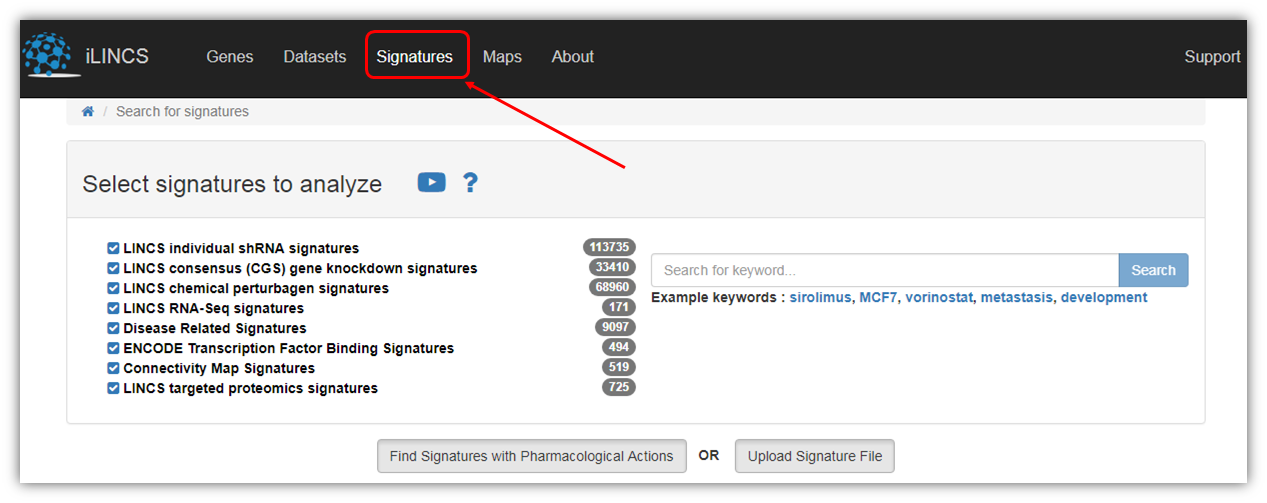
As shown in the figure above, there are 6 pre-computed LINCS signature libraries on iLINCS portal.
LINCS individual shRNA signatures - Transcriptional signatures of perturbations by short hairpin RNA based on L1000 assay. The signatures consist of differential gene expressions and p-values for 978 Landmark Genes measured by L1000 assay. The signatures were created by aggregating (ie averaging) Level 4 data for biological replicates as defined by the signatures metadata. Only signatures designated to be reproducible and self-connected ("gold") by the Broad institute are represented.
LINCS consensus (CGS) gene knockdown signatures - Transcriptional signatures were constructed by further aggregating signatures of individual shRNA perturbations. The signatures are based only on the 978 Landmark Genes measured directly by the L1000 assay.
LINCS chemical perturbagen signatures - Transcriptional signatures of perturbations by small molecules based on L1000 assay. Signatures were created by aggregating (ie averaging) Level 4 data for biological replicates as defined by the signatures metadata. Only signatures designated to be reproducible and self-connected ("gold") by the Broad institute are represented. The signatures consist of differential gene expressions and p-values for 978 Landmark Genes measured by L1000 assay.
LINCS RNA-seq signatures - Transcriptional signatures of baseline, unperturbed human breast cancer cells lines or cellular signatures for human cell lines for drug-induced toxicity caused by FDA approved drugs.
Disease Related Signatures - Transcriptional signatures constructed by comparing sample groups within the collection of public domain transcriptional dataset (GEO GDS collection). Each signature consists of differential expressions and associated p-values for all genes.
ENCODE Transcription Factor Binding Signatures - Transcription factor (TF) binding signatures constructed using ENCODE ChiP-seq data. Each signature consists of genome scale (ie for each gene) scores and probabilities of regulation by the given TF in the specific context (cell line and treatment). These signatures were developed using our in-house TREG methodology.
Connectivity Map Signatures - Transcriptional signatures of perturbagen activity constructed based on the version 2 of the original Connectivity Map dataset using Affymetrix expression arrays. Each signature consists of differential expressions and associated p-values for all genes when comparing perturbagen treated cell lines with appropriate controls.
LINCS targeted proteomics signatures - Transcriptional signatures of perturbagen activity constructed on the version 2 of the original Connectivity Map dataset using Affymetrix expression arrays. Each signature consists of differential expressions and associated p-values for all genes when comparing perturbagen treated cell lines with appropriate controls.
Search for LINCS signature
As seen in the figure below, one may search for LINCS signatures using a keyword of interest. Let's type-in "MCF7" in the search box as an example. All libraries are selected as a default option, but you may limit your search to a particular library of interest. Let's say you were interested only in precomputed signatures for LINCS chemical perturbagen signatures that include your search term, "MCF7". We will un-check all of the library selections, except for "LINCS chemical perturbagen signatures" and then click "Search".
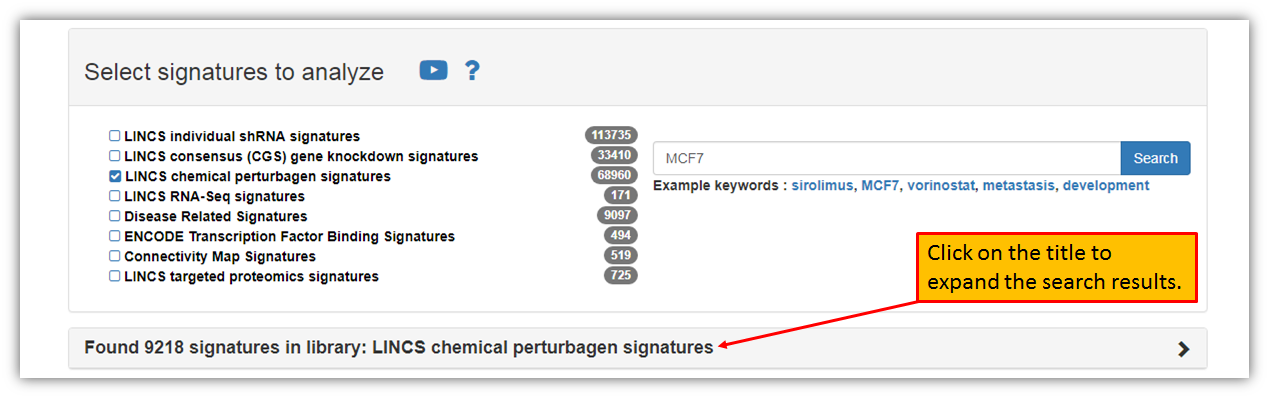
As seen in the figure above, using "MCF7" search term and searching only "LINCS chemical perturbagen signatures" have found over 9,000 signatures within a library. You may expand the search results by simply clicking on the search results as seen in the figure below.
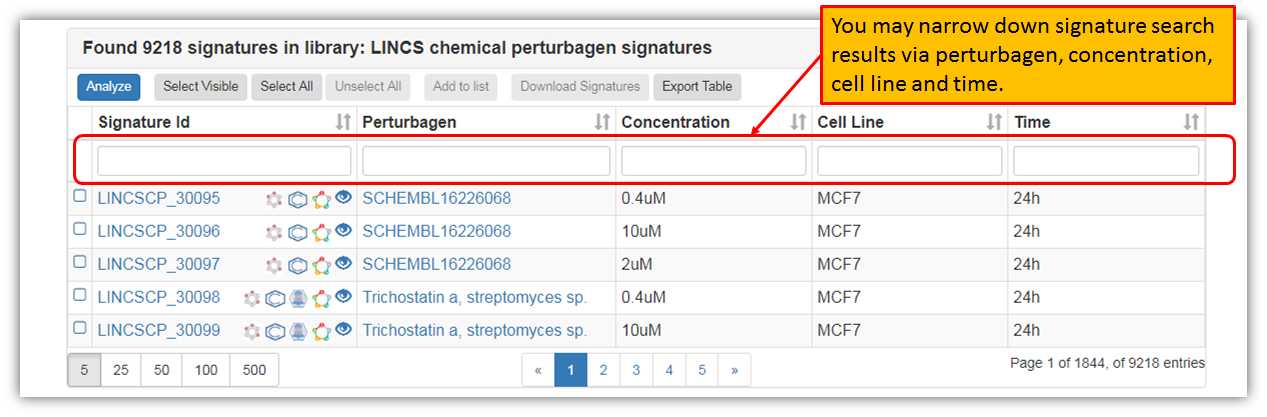
As seen in the figure above, you may browse through all the search results and you may also select a particular signature to view more details about it. Furthermore, you may narrow down signature search results in the search fields for a particular perturbagen or any other signature variable. You may also perform signature group analysis by selecting few signatures. Let's select first four signatures and click "Analyze" as shown in the figure below.
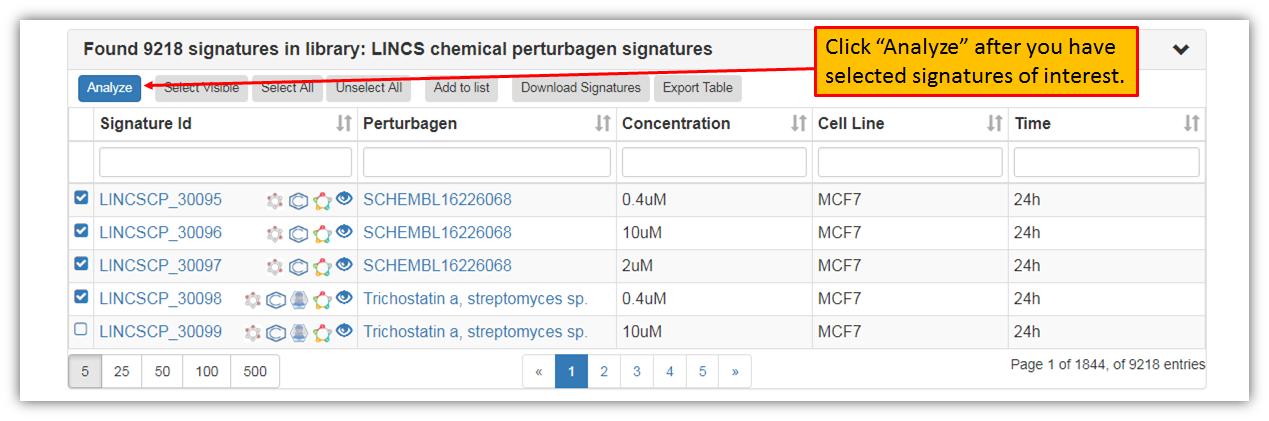
Clicking "Analyze" will provide you with a pop-up where you can confirm your selections. The default option of genes to be included in the analysis is 50, but you may increase or decrease this number to suit your preferences.
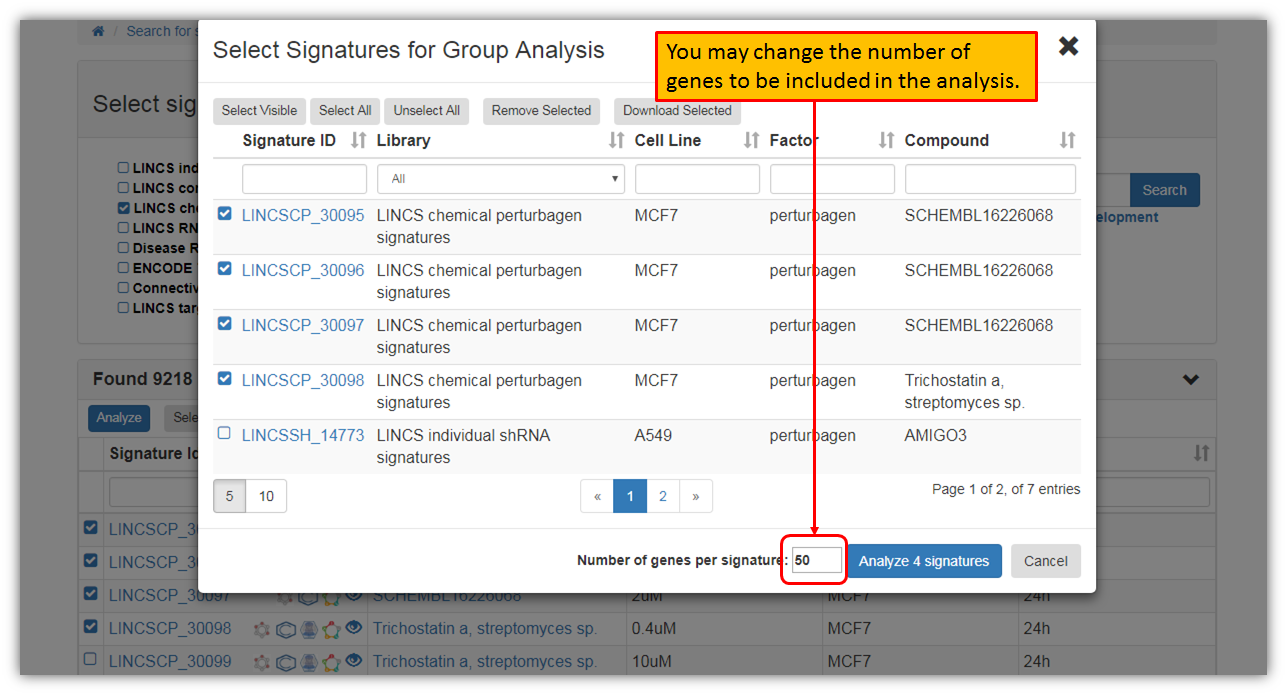
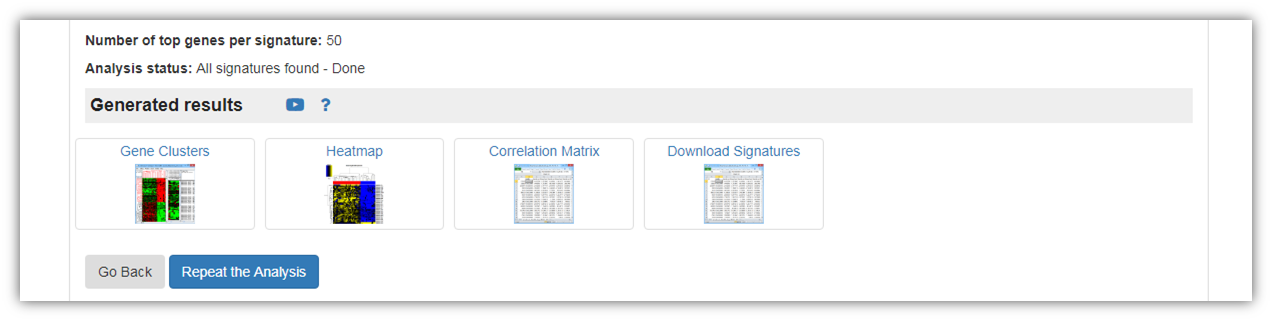
As seen in the figure below, you may view and/or download signature group analysis results.
Created with the Personal Edition of HelpNDoc: Free PDF documentation generator